Design and functionalisation of new nanodiscs
We design novel polymers and small amphiphilic compounds to improve membrane protein nanodiscs for biophysical and structural biology applications.
Amphiphilic copolymers have been introduced a few years ago as an alternative to traditional detergents. These polymers wrap around lipid bilayers like a belt, creating nanodiscs that entrap membrane proteins. We have established a diisobutylene/maleic acid (DIBMA) copolymer as a new protein-extracting compound that offers significant advantages: The ordering of lipid chains embedded in nanodiscs is only slightly affected by DIBMA and spectroscopic investigations of membrane proteins are also possible in the important ultraviolet spectral range. In addition, DIBMA tolerates elevated concentrations of divalent cations without precipitating, which is important for activity studies of many membrane proteins. We further developed a polymer derivative called Glyco-DIBMA that retains the same favorable properties but allows even more efficient solubilization of lipids and extraction of membrane proteins. Recently, we have shown that some small molecules, such as compounds with a diglucose head group, are able to extract membrane proteins and their surrounding lipids directly into nanodiscs, making them accessible for detailed studies.
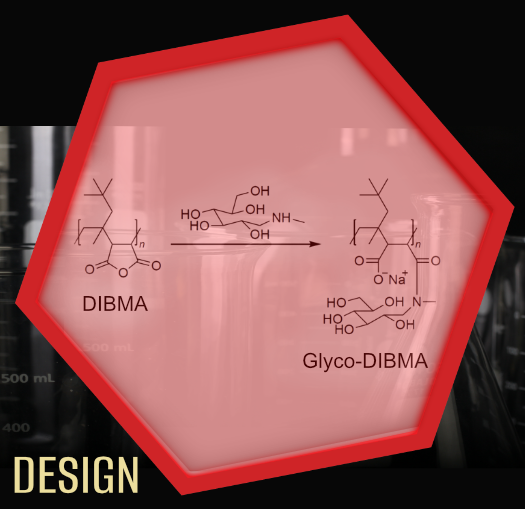
Image source background: pexels.com
ELMI microscopy image: Annette Meister
ONI microscopy image: David Glück
Biophysics of membrane proteins in nanodiscs
We are constantly refining biophysical methods to make them more compatible with nanodiscs and, conversely, we use the gained insights for the targeted optimization of nanodiscs.
As a second focus, we employ a wide range of biophysical methods to investigate nanodiscs and the membrane proteins and lipids embedded within them. Our repertoire of methods includes numerous spectroscopic, calorimetric and chromatographic techniques as well as light scattering and microfluidic diffusional sizing. In addition, high-resolution imaging techniques such as electron microscopy and single-molecule fluorescence measurements as well as high-throughput technologies such as mass spectrometry for proteomics and lipidomics are used. These complementary methods provide detailed information on the structure and size distribution of nanodiscs as well as on the conformation and dynamics of membrane proteins and lipids.
Nanodiscs as platforms for biomolecular applications
We are developing this nanodisc technology platform to elucidate the structures, dynamics, and interactions of membrane proteins with high resolution yet in an environment similar to the natural membrane.
A particularly important aspect of our interdisciplinary research is to use nanodiscs as versatile tools to answer biomolecular questions. Nanodiscs allow both the isolation of individual proteins and protein complexes as well as the extraction of entire protein libraries from different organisms and cells. This makes it possible to perform functional and interaction studies under laboratory conditions that can be well controlled but still mimic the natural environment of membrane proteins. For example, protein/protein interactions within a membrane environment can be studied without having to purify or reconstitute the membrane proteins for this purpose. Our nanodiscs are also used for high-resolution structural analyses of membrane proteins by electron microscopy, and can be used for ligand binding studies, thereby facilitating the identification of new drugs.

Image source background: pexels.com